Select your species of interest
Click on a species icon

Human
hg19
Homo sapiens
Chimpanzee
panTro4
Pan troglodytes
Mouse
mm10
Mus musculus
Rat
rn5
Rattus norvegicus
Chicken
galGal4
Gallus gallus
Frog
xenTro3
Xenopus tropicalis
Fruit fly
BDGP5
Drosophila melanogaster
Yeast
sacCer3
Saccharomyces cerevisiae
About R-loop
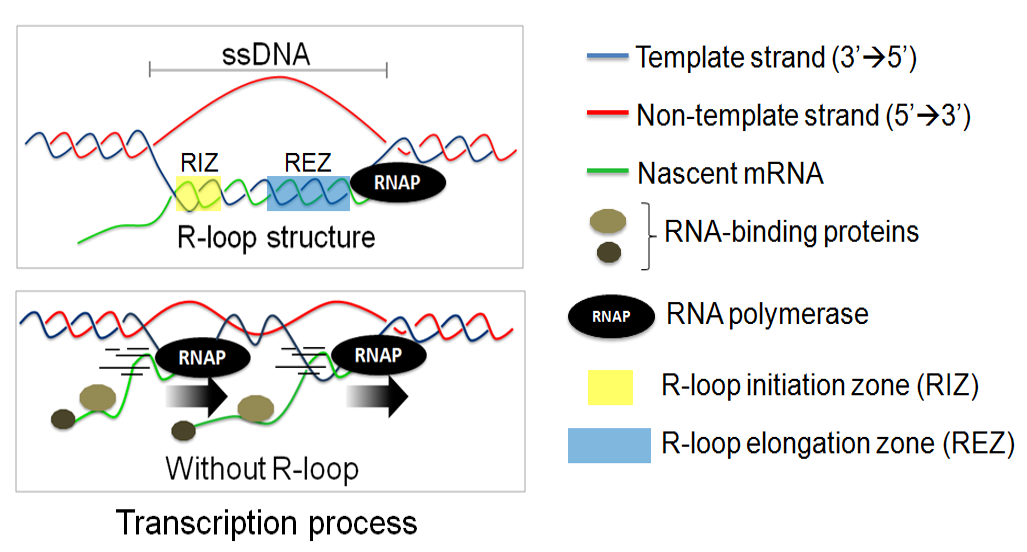 An R-loop is a three-stranded nucleic acid structure,
formed during transcription, which comprises of nascent RNA hybridized with the DNA template, leaving the
non-template DNA single-stranded. more... (see Figure on right panel)
An R-loop is a three-stranded nucleic acid structure,
formed during transcription, which comprises of nascent RNA hybridized with the DNA template, leaving the
non-template DNA single-stranded. more... (see Figure on right panel)
R-loop is often formed during transcription and its formation has been observed for several species. Their roles in the regulation of transcription, splicing, telomere maintenance, genome instability, mutagenesis, cell proliferation and differentiation as well as diseases involvement have been demonstrated.
R-loop DB was originally constructed as a collection of R-loop forming sequences (RLFS) predicted computationally in the human genome based on our quantitative model of RLFS (QmRLFS). Here, our R-loop DB additionally includes chromosome coordinates and annotation of many hundred thousand of newly predicted RLFSs computationally defined in the genomes of eight species. These RLFS were predicted by an extended version of the QmRLFS model which can produce more accuracy of the prediction results. Genome-wide experimentally detected RNA-DNA hybrid data were also integrated into the R-loop DB.
Essentially, we have updated the R-loop DB with the extension and specification of the collection of RLFS in non-coding and protein-coding genes, promoter and downstream gene regions. Further comprehensive graphical tools are also developed for multiple annotation track visualization and analysis of the RLFS loci.